Bengaluru: Genetic engineering researchers have discovered a powerful tool that can be used to edit genes on a larger scale. This tool will allow researchers to rearrange, recombine, invert, duplicate, move, and perform other editing operations on very long DNA sequences. In the future, this is expected to lead to more advanced gene editing therapeutics and treatments for diseases.
This next generation genomic design method, called the bridge recombinase mechanism, exists naturally in our genetic machinery, and can be used to program and edit DNA.
This gene editing method, which exists naturally and has now been discovered, enhances the human ability to edit genomes beyond the capabilities and scope of CRISPR (clustered regularly interspaced short palindromic repeats), a technology that can be used to modify the DNA of living organisms. It utilises mobile genetic elements or “jumping genes”, which cut and paste themselves into genomes and are present in all forms of life, performing on-the-go DNA manipulation through all living beings.
The findings are reported in the form of two papers characterising the discovery and the working of a ‘bridge’ RNA molecule which can then be reprogrammed as needed, and the structural mechanism behind the discovered recombination ability of these genes. Both papers were published June 26 in the journal Nature. The study announcing the discovery was led by Matthew G. Durrant of the University of California, Berkeley, and the one describing the structural mechanism of the recombination was led by Masahiro Hiraizumi of the University of Tokyo.
Also read: Pune astronomers identify galaxy in Milky Way’s neighbourhood as ‘explosive factory’ of gamma rays
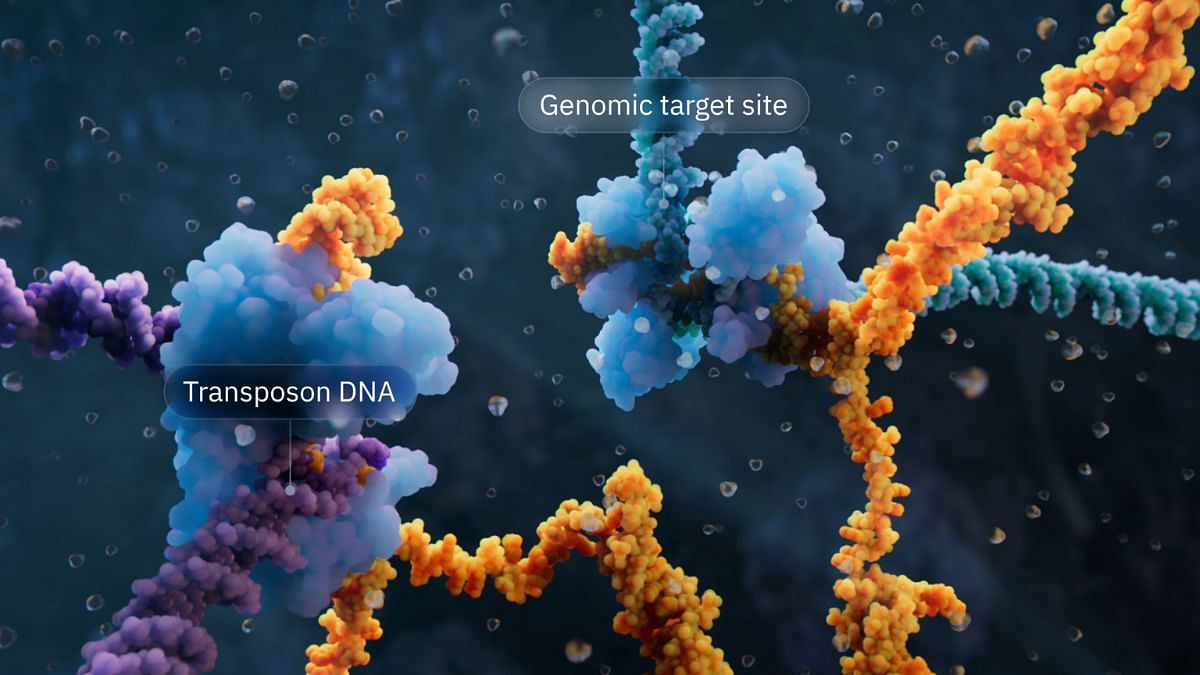
Jumping genes, and how they act as bridges
Jumping genes are minimal segments of DNA that have the recombinase enzyme, which binds this DNA to other DNA, along with extra DNA segments at the ends of the genes. The team discovered that these bits of extra DNA at the ends of jumping genes get joined together and convert the DNA double helix structure into a single-stranded RNA molecule that folds into two loops.
This can then bind to two sets of DNA, the donor and the target, with each loop of the element binding independently to the donor segment and target segment. The target DNA segment is the one that needs to be modified, and the donor segment is the one whose parts will be used to modify the target sequence. Thus, this jumping gene then functions as a bridge that recombines two bits of unconnected DNA.
The researchers also made the important discovery that the donor loop and the target loop can be programmed independently, offering great flexibility in inserting or recombining sequences to DNA.
The name of the jumping gene is IS110, which stands for Insertion Sequence, and such sequences are found in ample quantities in bacteria, which were used for the study (E. coli). They roam around the body, cutting and pasting themselves, repairing DNA and modifying it daily.
“The IS110 bridge recombination system expands the diversity of nucleic-acid-guided systems beyond CRISPR and RNA interference, offering a unified mechanism for the three fundamental DNA rearrangements — insertion, excision and inversion — that are required for genome design,” write the authors on the paper.
DNA insertion is a genetic process in which a segment of DNA is added to a different DNA segment, excision is a mechanism in which a damaged DNA segment is removed, and inversion is a method in which a piece of DNA in a chromosome gets reversed.
The role of the flanking DNA (sequences found on either side of the DNA fragment of interest) present around the recombinase genes of these mobile sequences has been understood for the first time. One of the authors on the paper compared bridge RNA to a universal power adapter that makes the IS110 compatible with any outlet or DNA segment.
The second paper outlines the mechanism reports results from cryo-electron microscopy (a technique in which samples are cooled to cryogenic temperatures to determine three-dimensional structures of internal fragments), which indicate that the recombination occurs in three steps.
Also read: IIT-Madras, NASA jointly study multidrug-resistant pathogens on International Space Station
How RNA bridge differs from CRISPR, RNAi
Both CRISPR and RNA Interface (RNAi), another form of gene editing, work by blocking gene expression or activating it. RNAi does so by targeting RNA molecules, which then do the work of editing genes, while CRISPR edits DNA directly.
The RNA bridge, on the other hand, modifies large sequences of DNA instead of just a tiny segment consisting of a handful of genes. This greatly expands the scope of changes that can be brought about by the technique, which is also a limitation of the method.
This is because the technique modifying large chunks of DNA sequences increases the risk of unintentional consequences.
A limitation of the findings is that the studies were performed in vitro or in the lab on bacteria. To apply to any humans, they would need to be tried on animal models, and specifically mammal models, first.
(Edited by Radifah Kabir)
Also read: Life-sustaining freshwater first appeared on Earth 4 billion years ago, finds Nature study