Keeping the genetic information stored in genomic DNA intact during the cell division cycle is crucial for almost all lifeforms. Extensive DNA damage invariably causes various adverse genomic rearrangements, which can lead to cell death in the best cases and to the occurrence of diseases like cancer in the worst cases.
Fortunately, cells in all three domains of life share a peculiar error-free mechanism for maintaining genetic information, known as homologous recombination (HR).
The process of HR starts when a cell encounters DNA damage during DNA synthesis or afterwards, initiating a cascade of events. The damaged DNA is first resected or cut to create single-stranded ends near the damaged site. These ends are then matched to their corresponding region in an available replicated chromosome, also known as “sister chromatid,” which is essentially used as a template to repair the damaged DNA.
As one might expect, the HR pathway involves a myriad of proteins and cellular machinery. While most of these proteins and cellular machinery are well-studied, some of them remain somewhat enigmatic. Such is the case of the regulators of RAD51, a protein responsible for repairing DNA double-strand breaks.
Normally, RAD51 forms filaments that help preserve DNA replication forks—transient arrangements of DNA that often occur during DNA replication, such as in replication fork collapse. Proper regulation of RAD51, as well as the degradation of these filaments after their purpose has been served, is essential for HR.
However, the precise mechanisms by which abnormal RAD51 accumulation leads to genetic instability are not completely understood, and many positive and negative RAD51 regulators remain obscure.
Now, however, in a recent article published in Nucleic Acid Research on 10 April 2024, a research team led by Professor Miki Shinoara from the Department of Advanced Bioscience, Kindai University, Japan, investigated the close relationship between RAD51 and FIGNL1, one of its key regulators. The study was co-authored by Kenichiro Matsuzaki, also from the Department of Advanced Bioscience, Kindai University, and sheds some much-needed light on the intricacies of the HR process.
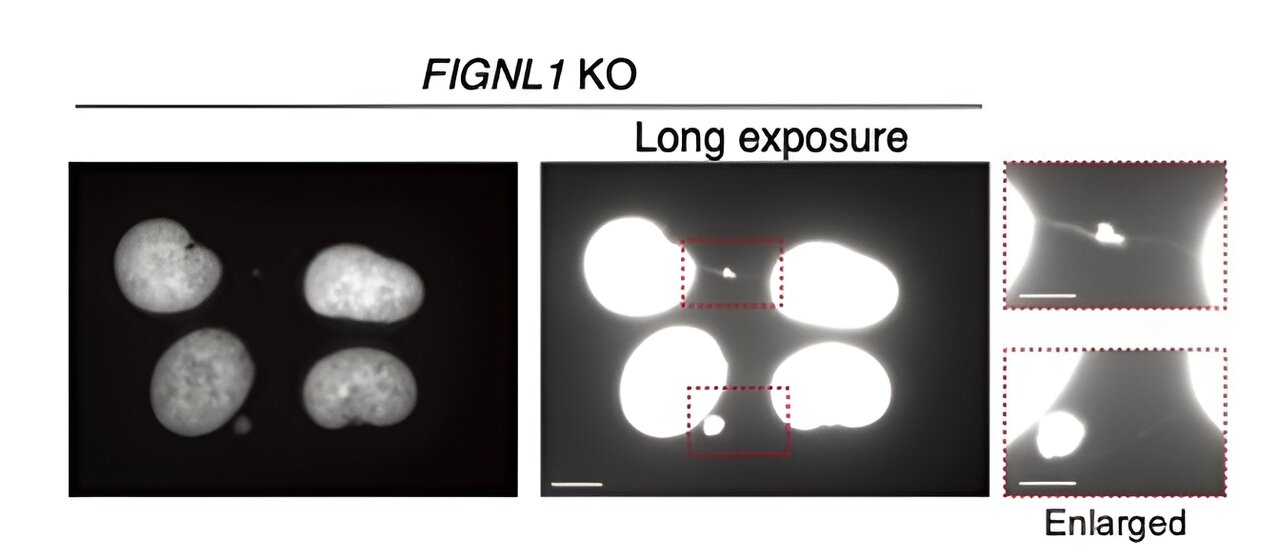
First, the researchers genetically engineered human cells that did not express FIGNL1 (that is, FIGNL1 KO cells), using the well-established CRISPR/Cas9 method. Then, using advanced immunostaining techniques involving carefully selected antibodies and fluorescence microscopy, they visualized the HR process in detail, looking for indicators of abnormalities.
By combining this approach with a plethora of other experimental procedures, such as western blotting, cell cycle analysis, protein assays, and genomic and transcriptomic analyses, they managed to get a comprehensive picture of what happens in a cell when FIGNL1 is missing.
The results reveal that FIGNL1 is a highly specialized RAD51-dismantling enzyme that is necessary for proper chromosome separation after replication forks are “disassembled.”
More specifically, when RAD51 filaments are not fully dismantled, abnormal events occur during mitosis that produce unresolved intermediates. This ultimately leads to the formation of so-called ‘chromosome bridges’ between the sister chromatids. These ultra-fine structures are very detrimental to the normal operation of the cell, causing the propagation of catastrophic genetic information.
Understanding the finer details of the HR pathway, its key players, and its many sub-processes is extremely important not only from a biological perspective, but also from a medical standpoint.
“Cell death due to dysregulation of HR is an important mechanism by which anticancer drugs exhibit cancer cell-specific cytotoxicity,” explains Prof. Shinohara. “Until now, the main target has been HR activation deficiency, but the results of this study show that persistent activation of RAD51 also exhibits cytotoxicity and can be a molecular target for anticancer drugs.”
Moreover, the cellular machinery involved in the HR pathway can be leveraged as a powerful bioengineering tool.
“HR is a well-conserved system among most species and is also tightly connected to gene modification technologies, such as genome editing and gene targeting technologies,” comments Prof. Shinohara, “Thus, elucidating the mechanisms that control recombinase activity, such as that of RAD51, may contribute to increasing the efficiency of gene modification techniques.”
Worth noting, genetic engineering is a highly effective avenue for increasing crop yield and for customizing microbial organisms for tasks such as bioremediation, which addresses various modern world problems.
Overall, the findings of this study not only shed light on a universal biological process but also pave the way toward a better understanding of cellular mechanisms for important drug discoveries and progress in the field of genetic engineering.
More information:
Kenichiro Matsuzaki et al, Human AAA+ ATPase FIGNL1 suppresses RAD51-mediated ultra-fine bridge formation, Nucleic Acids Research (2024). DOI: 10.1093/nar/gkae263
Provided by
Kindai University
Citation:
Researchers study the intricacies of homologous recombination and abnormal chromosome bridges (2024, May 7)
retrieved 8 May 2024
from https://phys.org/news/2024-05-intricacies-homologous-recombination-abnormal-chromosome.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.